2.2. Project: Femur Fitting¶
This project was created as part of the Computational Physiology module in the MedTech CoRE Doctoral Training Programme.
This project requires you to put together what you have learned in the tutorials to define a complete workflow which will create a customised model of the end of a femur bone by fitting it to medical images, perform some computational simulation with it and visualise the result.
2.2.1. Outline¶
You will need to start MAP Client and create a new workflow via the menu item . This just requires you to select a folder: create a new, empty folder, for example “femurproject” on the Desktop, and select it.
At this point the MAP Client interface will show a blank project and a list of plugins you can use, as shown in Fig. 2.10:
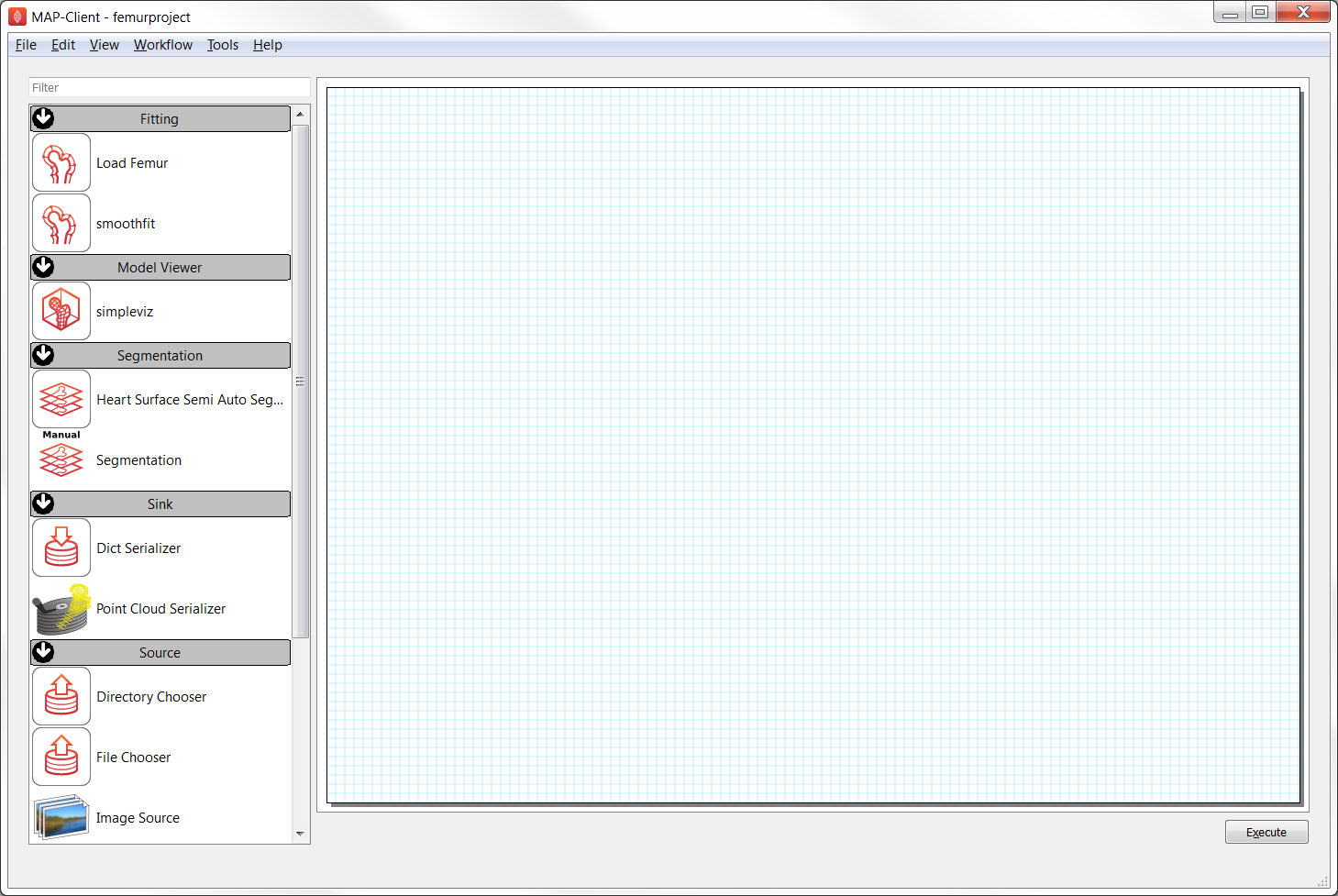
Fig. 2.10 The MAP Client interface showing plugins to use for the femur project. (Note that your version may have additional plugins.)
For this project you will use the following plugins for steps in the workflow:
- Image Source - for selecting a folder of images to digitise
- Segmentation (Manual) - for digitising points in the image stack for segmenting the femur surface
- File Chooser - for configuring the input model to fit
- smoothfit - for fitting the input model to the digitised points
- Load Femur - for performing some heavy math on the fitted model
- simpleviz - for visualising the results
These are added to your workflow by dragging them into the working area (with the blue grid, like graph paper). Each step must be configured by clicking on its green, gear-shaped settings icon, to give it a unique name in this workflow (intermediate data is saved under this name), and set any other options for sources to the workflow. You will need to connect the input and output ports for each step.
Tips for completing the project:
- For initial input to the Segmentation tool you will want to select folder
DTP-data/complete_workflow/knee_mri(the location of theDTP-datafolder will be given to you). - When digitising points on the medical images, make sure you create adequate points to define the curved surfaces: it’s easy to underspecify the shape at the end of rounded body. You may wish to rotate the plane you’re digitising on to suit capturing the femur shape. It takes a while to digitise so routinely save the data, and a final time before clicking ‘done’. Next time you run the workflow you can click ‘load’ to reload it.
- For input to the smoothfit tool you’ll need an initial femur model. The suggested model is
DTP-data/project-data/surfacefit-output-model3.exfile(which was itself the output of a fit!). - The smoothfit step has 3 inputs: the top one is the initial model, the bottom one is a cloud of data points (as output by the Segmentation tool). The middle one is unused here as it used as an alternative data point cloud, read from an EX format file. Be sure to save your final transformation so you can re-load it next time the workflow is run.
- Note it takes a few seconds to perform each fit iteration, and a similar amount of time to perform the Load Femur maths.
- When initially visualising you will see nothing! Create surfaces and try to view the stress on it. Add a colour bar and try creating some other graphics. Save the resulting image.
Your final visualisation will look something like Fig. 2.11:
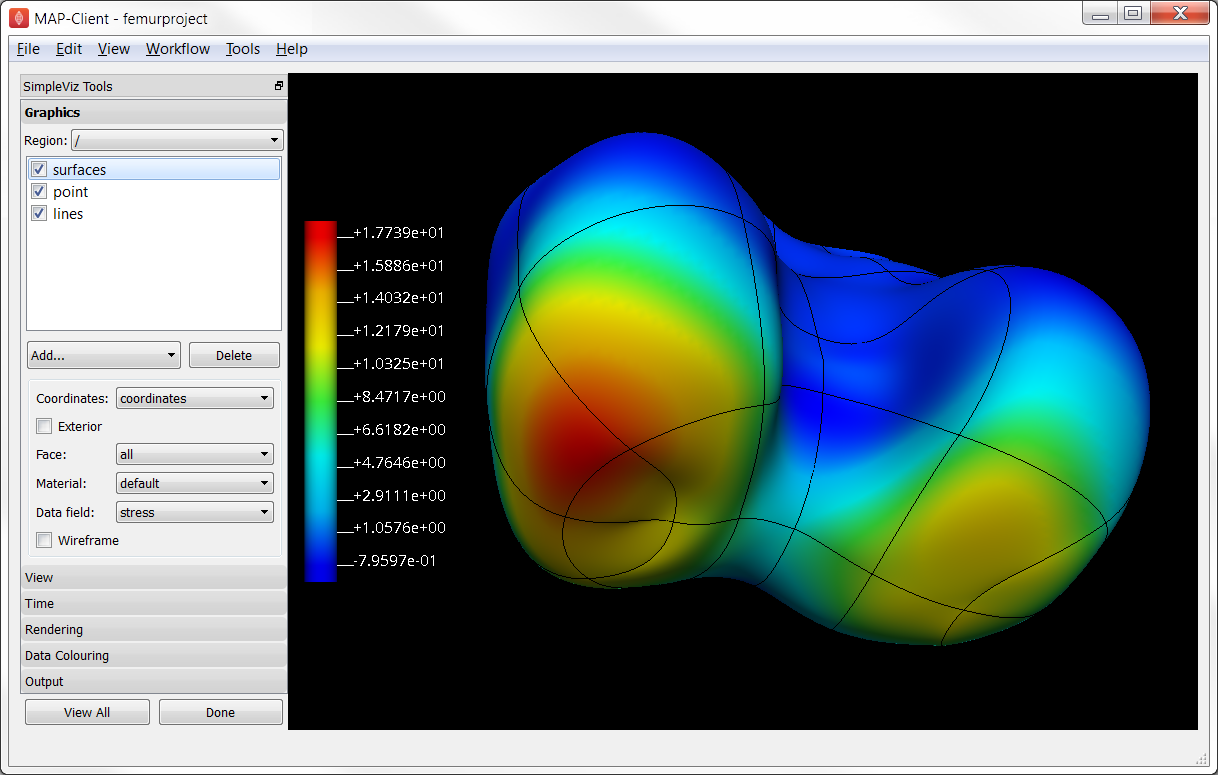
Fig. 2.11 Visualisation of the final result of the femur project. Your result may use different graphics, and will differ because your digitisation and fitting will always make a unique result.
